Retinotopic Mapping Tutorial
A tutorial on processing MRI data from a retinotopic mapping experiment.
Table of Contents
- Introduction
- Stimulus and Scan
- FreeSurfer
- Preprocessing the EPIs
- pRF Models
- Surfaces and Parameter Visualization
- Atlas Generation
- Bayesian Inference of Retinotopic Maps
Introduction
Retinotopic mapping experiments are very common in visual fMRI research and are performed frequently enough in the Winawer lab that we have developed a rough pipeline for their preparation and analysis. This document will step through this pipeline using a dataset collected at NYU, demonstrating the various processing done at each stage. This document is written primarily for use with NYU’s Center for Brain Imaging (CBI). Details about the preprocessing steps performed by CBI for NYU researchers (e.g., B0 fieldmap correction) can be found on the CBI intranet. This tutorial assumes that you have already downloaded your data from Tesla (non-NYU users will need to perform some inference in figuring out how to adapt the first few steps to their own local setups).
Stimulus and Scan
The stimulus and scan protocol used in the experiment that is analyzed in this document was nearly identical to that used in the Human Connectome Project (HPC); see their page on protocols for more information. In the particular experiment analyzed here, we performed 8 runs of retinotopic mapping, each of which lasted 192 TRs (1 second each). We additionally collected 1 T1-weighted image at a resolution of 0.8 mm\(^3\).
This tutorial begins in my ~/Downloads/rscan_20180205 directory (using Mac OS 10.13.2) to which I
have already downloaded all scan files from the CBI’s Tesla file-server. For
information on how to do this, wee the related page on the Winawer-lab
wiki. At the start of the
analyses detailed below, I observed the following directory contents:
> pwd
/Users/nben/Downloads/rscan_20180205
> ls
01+AAHead_Scout_64ch-head-coil 16+cmrr_mbepi_s6_2mm_66sl_PA_TR1.0_SBRef
02+AAHead_Scout_64ch-head-coil_MPR_sag 17+cmrr_mbepi_s6_2mm_66sl_PA_TR1.0
03+AAHead_Scout_64ch-head-coil_MPR_cor 18+cmrr_mbepi_s6_2mm_66sl_PA_TR1.0_SBRef
04+AAHead_Scout_64ch-head-coil_MPR_tra 19+cmrr_mbepi_s6_2mm_66sl_PA_TR1.0
05+FMRI_DISTORTION_AP_2mm_66sl 20+cmrr_mbepi_s6_2mm_66sl_PA_TR1.0_SBRef
06+FMRI_DISTORTION_PA_2mm_66sl 21+cmrr_mbepi_s6_2mm_66sl_PA_TR1.0
07+AAHead_Scout_64ch-head-coil 22+cmrr_mbepi_s6_2mm_66sl_PA_TR1.0_SBRef
08+AAHead_Scout_64ch-head-coil_MPR_sag 23+cmrr_mbepi_s6_2mm_66sl_PA_TR1.0
09+AAHead_Scout_64ch-head-coil_MPR_cor 24+cmrr_mbepi_s6_2mm_66sl_PA_TR1.0_SBRef
10+AAHead_Scout_64ch-head-coil_MPR_tra 25+cmrr_mbepi_s6_2mm_66sl_PA_TR1.0
11+FMRI_DISTORTION_AP_2mm_66sl 26+cmrr_mbepi_s6_2mm_66sl_PA_TR1.0_SBRef
12+FMRI_DISTORTION_PA_2mm_66sl 27+cmrr_mbepi_s6_2mm_66sl_PA_TR1.0
13+T1_MPRAGE 28+cmrr_mbepi_s6_2mm_66sl_PA_TR1.0_SBRef
14+cmrr_mbepi_s6_2mm_66sl_PA_TR1.0_SBRef 29+cmrr_mbepi_s6_2mm_66sl_PA_TR1.0
15+cmrr_mbepi_s6_2mm_66sl_PA_TR1.0 99+PhoenixDocument
For those not familiar with the CBI’s naming convention, each of these items listed above is a directory, and each contains two files: a text file whose contents are a reproduction of the original DICOM header for the scan, and a NifTI file whose contents are the actual 3D or 4D scan measurements. The directories containing the scans are numbered by the order in which they were collected.
Note. Looking closely at the above directory contents, one might notice that we performed multiple scout and distortion scans prior to performing a T1 and the 8 retinotopy scans. This was, in fact, the result of a problem getting our stimulus to display propertly. In an ideal world this would never happen, but in reality, it is quite common to have a scan or two that must be discarded or ignored in a study. In this case, we will discard scans 1-6 (and note that we will not use the scout scans 7-10 in these analyses). The commands we use below will reflect the fact that we are ignoring these scans.
FreeSurfer
FreeSurfer is a software suite for processing anatomical MRI data; it includes a large number of useful algorithms that, among other things, identify the brain, strip the skull, identify white- and gray-matter voxels, perform various normalizations, tesselate the white and pial surfaces, and perform cortical surface alignment to an average anatomical atlas.
recon-all
The first step in processing a retinotopy experiment is generally to process the T1-weighted
anatomical image. This step is highly complicated, but that complication is entirely performed by
FreeSurfer, using a single command, recon-all.
> ls 13+T1_MPRAGE/
wl_subj046_ColorRetinotopy+13+T1_MPRAGE-header.txt wl_subj046_ColorRetinotopy+13+T1_MPRAGE.nii
> recon-all -subjid wl_subj046 -i 13+T1_MPRAGE/wl_subj046_ColorRetinotopy+13+T1_MPRAGE.nii -all &> ./wl_subj046_recon-all.log
Estimated Runtime: 3-12 hours, depending on your computer, FreeSurfer version, the subject, and the scan. I have never made a habit of timing this command and usually just assume that it will run overnight. I have heard that subjects with more curvature in their cortices require more time to process (which makes some intuitive sense given that FreeSurfer performs cortical surface tesselation), but I have never confirmed this myself.
In the above command, we call FreeSurfer’s recon-all command, which handles the importing of
subjects into FreeSurfer’s subject directory by performing an enormous suite of processing on the
T1-weighted anatomical image. Documentation on FreeSurfer is not always up-to-date and
comprehensive, but official information about the recon-all process is available
here. In my experience, more can usually be
learned from the recon-all dev-table,
though understanding the steps documented there may require requesting --help documentation from
the various FreeSurfer commands. For example, to understand the normalization procedure used in the
first stage of recon-all, note that the command mri_normalize is used; the best information about
this processing step will likely come from running mri_normalize --help.
Checking the Results
Although FreeSurfer’s results are generally fine, it’s not a bad idea to check for obvious problems
in the resulting data files. An easy way to do this is to use the program freeview, which comes
with FreeSurfer. Freeview itself has variety of visualization functions that it performs, the extent
of which is beyond the scope of this document. This
webpage
gives a brief tutorial and links to other resources, however.
To check our subject, we will first look at their brain.mgz file to make sure it looks reasonable,
then overlay on it the ribbon.mgz file to make sure that there aren’t major errors in the
segmentation. To start this, we can run freeview from the command-line:
# switch to the subject's new FreeSurfer directory
> cd /Volumes/server/Freesurfer_subjects/wl_subj046
> ls mri/
T1.mgz orig
aparc+aseg.mgz orig.mgz
aparc.a2009s+aseg.mgz orig_nu.log
aseg.auto.mgz orig_nu.mgz
aseg.auto_noCCseg.label_intensities.txt rawavg.mgz
aseg.auto_noCCseg.mgz rh.ribbon.mgz
aseg.mgz ribbon.mgz
brain.finalsurfs.mgz segment.dat
nu.mgztalairach.label_intensities.txt talairach.log
brainmask.auto.mgz talairach_with_skull.log
brainmask.mgz talairach_with_skull_2.log
ctrl_pts.mgz transforms
filled.mgz wm.asegedit.mgz
lh.ribbon.mgz wm.mgz
mri_nu_correct.mni.log wm.seg.mgz
mri_nu_correct.mni.log.bak wmparc.mgz
norm.mgz brain.mgz
nu_noneck.mgz
# Start Freeview; load the brain.mgz file
> freeview -v mri/brain.mgz
The above command should open a window that looks like this:
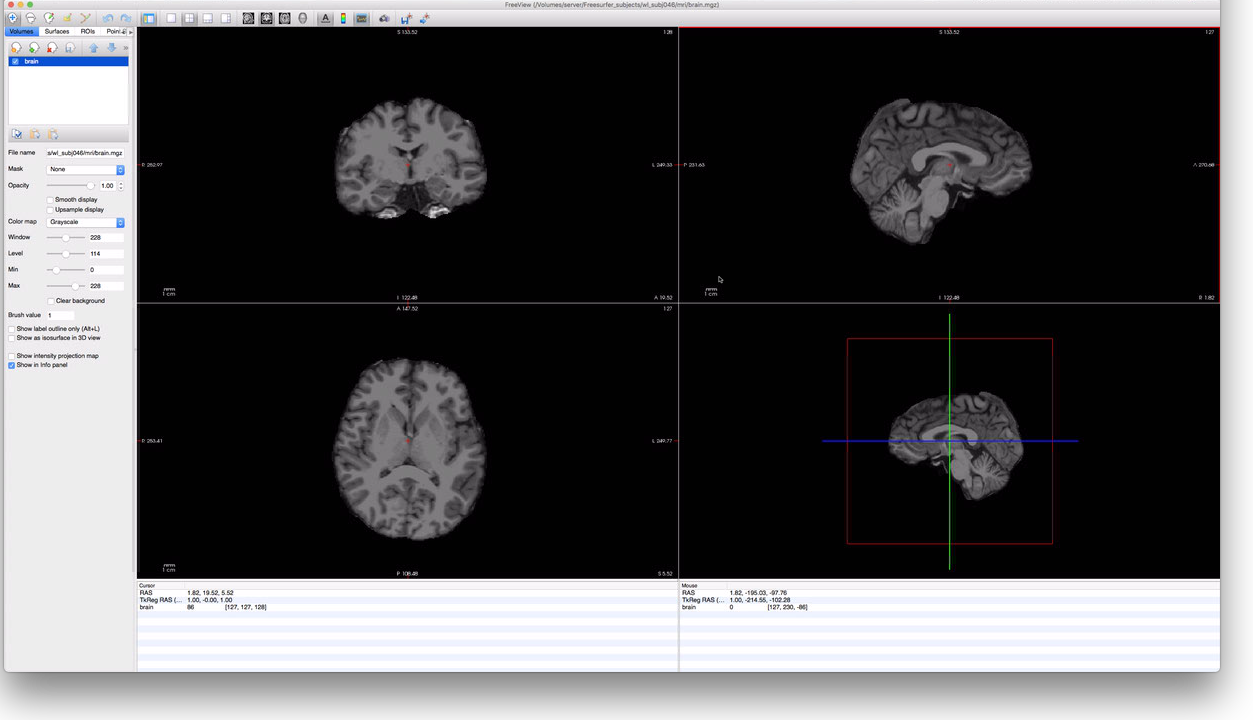
By scrolling through this subject’s data, it should be easy to determine if FreeSurfer correctly
identified the location of the subject’s brain; in this case, it did a pretty good job. To check,
the segmentation, however, we must load the ribbon file. The ribbon contains the same voxels labeled
as either LH white-matter, LH gray-matter, RH white-matter, RH gray-matter, or none. If we click on
the “Load Volumes” button in the upper left corner of the FreeView window (the head with the green
plus icon), we can add a volume to the display. Click this button then select the same subject’s
ribbon.mgz file, also in the mri/ directory. At the bottom of the import-file window is a list
of options for the display/color for the volume; select “Lookup Table” for this. Once you’ve done
this, you should have a FreeView window that looks something like this:
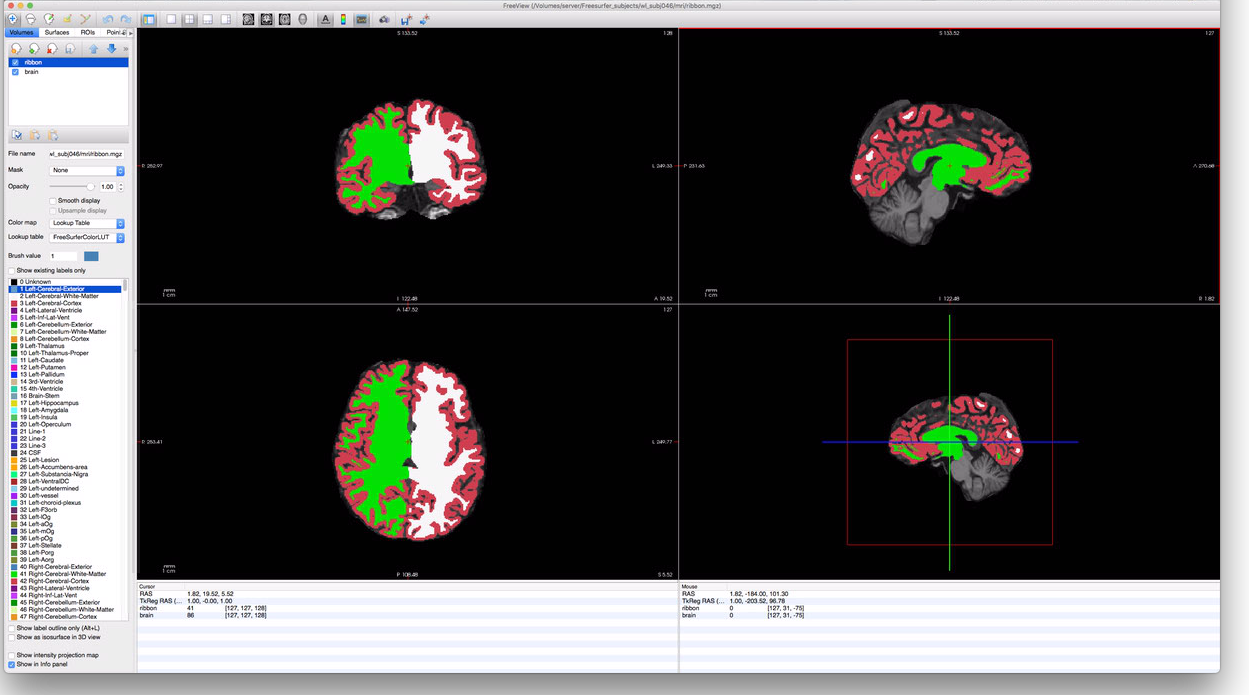
To make the ribbon more transparent, you can adjust the “Opacity” slider in the menu-bar on the
left. Additionally, you can check or uncheck the “ribbon” volume near the upper-left corner to
toggle display of the ribbon file altogether. It’s generally a good idea to look through the
subject’s occipital cortex to make sure that the segmentation assigned by the ribbon looks
reasonable with respect to the voxels in the brain.mgz; segmentation errors in the ribbon can
result in significant errors during the cortical surface generation. Although repairing errors is
beyond the scope of this tutorial, it involves hand-editing the wm.mgz file and restarting
recon-all at an intermediate stage. The wm.mgz file can be hand-edited using
ITK-Snap, and this
page gives an
introduction to processing such repairs.
Finally, we can check that the cortical surfaces were constructed reasonably. To do this, we click
on the Surfaces tab in the upper-left corner of FreeView, then on the “Load Surface” icon (the
brain with the green plus in the upper left). This will bring up a surface-loading dialog-box in
which you can navigate to your subjects surf/ directory (in their FreeSurfer subject directory)
and select the lh.white surface file. This will yield a display that looks something like this:
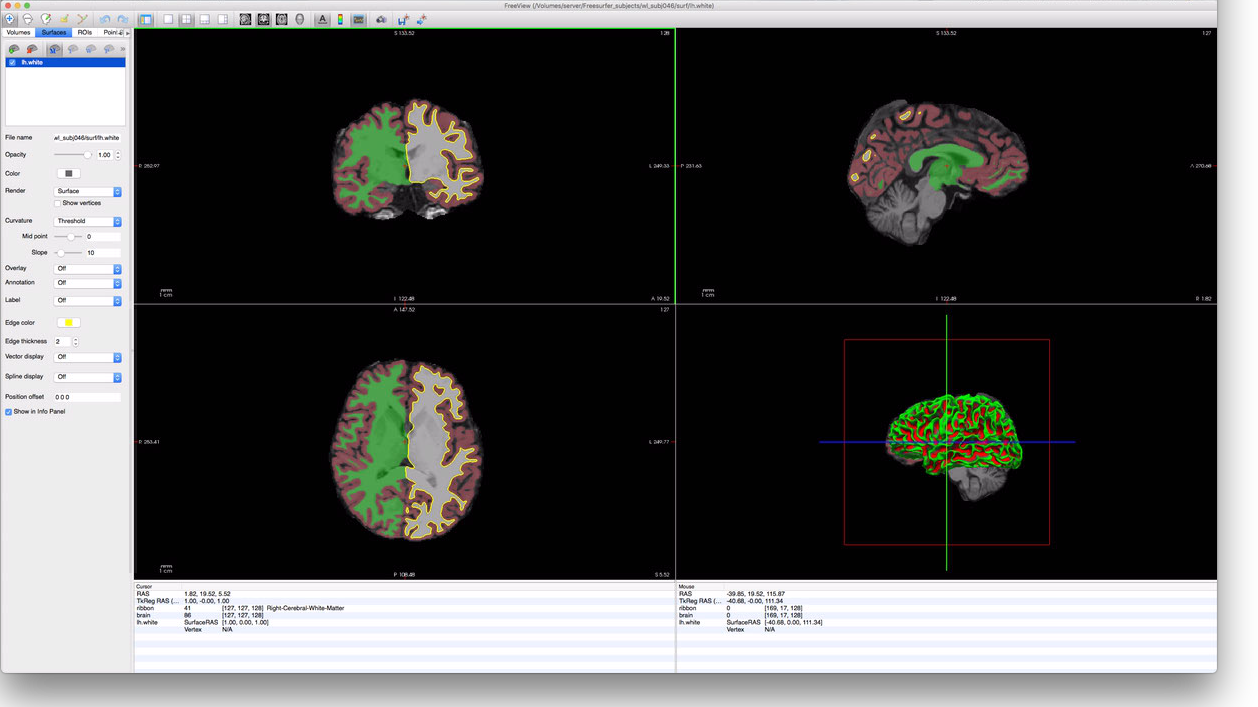
Again, repair of these surfaces is beyond the scope of this document; though, in the author’s
experience, most cortical surface errors are due to segmentation errors and should be corrected
prior to surface generation. Other surface files that should be checked for errors are rh.white,
lh.pial, and rh.pial.
Preprocessing the EPIs
The next step, after processing the anatomical scan, is to preprocess the EPI scans. Most of this step is performed by the prisma_preprocess.py script; though the final step is performed by the to_freesurfer.py script.
prisma_preproc.py
The prisma_preproc.py script does the bulk of the preprocessing work for EPIs. The script, by
Serra Favila, encapsulates a number of complicated steps including the unwarping/undistorting of the
EPI images, motion correction, and calculation of the alignment to the subject’s
FreeSurfer-processed anatomy data. This script can be found at the Winawer lab github
page in the MRI_tools
repository. Detailed information about running prisma_preproc.py can be found on this Winawer lab
wiki page that documents it.
Before running the prisma_preproc.py script, it is worthwhile to setup a VistaSoft session
directory; for retinotopy, we typically keep our retinotopy sessions in the Projects/Retinotopy
directory on our lab webserver. This bash session demonstrates this setup:
# Make the subject directory...
> mkdir /Volumes/server/Projects/Retinotopy/wl_subj046
> cd /Volumes/server/Projects/Retinotopy/wl_subj046
# ...and the session directory...
> mkdir 20180205_ColorRetinotopy
> cd 20180205_ColorRetinotopy
# Make the various directories
> mkdir Raw
> mkdir Preproc
> mkdir Stimuli
> mkdir Outputs
> mkdir Code
# Copy over the prisma_preproc.py and to_freesurfer.py scripts;
# the ~/Code/MRI_tools directory contains the MRI_tools repo.
> cp ~/Code/MRI_tools/preprocessing/*.py Code/
# We will also want the retinotopy code later.
> cp ~/Code/MRI_tools/retinotopy/*.py Code/
# Also copy over stimulus movie and parameter files; these
# specific files are for the experiment we ran:
> cp /Volumes/server/Projects/Retinotopy/ColorRetinotopyShared/stimuli/scan_*.mat Stimuli
# Transfer over the raw data
> mv ~/Downloads/rscan_20180205/* Raw/
After executing the above, we are ready to run the preprocessing script:
> cd Raw
> ls
01+AAHead_Scout_64ch-head-coil 16+cmrr_mbepi_s6_2mm_66sl_PA_TR1.0_SBRef
02+AAHead_Scout_64ch-head-coil_MPR_sag 17+cmrr_mbepi_s6_2mm_66sl_PA_TR1.0
03+AAHead_Scout_64ch-head-coil_MPR_cor 18+cmrr_mbepi_s6_2mm_66sl_PA_TR1.0_SBRef
04+AAHead_Scout_64ch-head-coil_MPR_tra 19+cmrr_mbepi_s6_2mm_66sl_PA_TR1.0
05+FMRI_DISTORTION_AP_2mm_66sl 20+cmrr_mbepi_s6_2mm_66sl_PA_TR1.0_SBRef
06+FMRI_DISTORTION_PA_2mm_66sl 21+cmrr_mbepi_s6_2mm_66sl_PA_TR1.0
07+AAHead_Scout_64ch-head-coil 22+cmrr_mbepi_s6_2mm_66sl_PA_TR1.0_SBRef
08+AAHead_Scout_64ch-head-coil_MPR_sag 23+cmrr_mbepi_s6_2mm_66sl_PA_TR1.0
09+AAHead_Scout_64ch-head-coil_MPR_cor 24+cmrr_mbepi_s6_2mm_66sl_PA_TR1.0_SBRef
10+AAHead_Scout_64ch-head-coil_MPR_tra 25+cmrr_mbepi_s6_2mm_66sl_PA_TR1.0
11+FMRI_DISTORTION_AP_2mm_66sl 26+cmrr_mbepi_s6_2mm_66sl_PA_TR1.0_SBRef
12+FMRI_DISTORTION_PA_2mm_66sl 27+cmrr_mbepi_s6_2mm_66sl_PA_TR1.0
13+T1_MPRAGE 28+cmrr_mbepi_s6_2mm_66sl_PA_TR1.0_SBRef
14+cmrr_mbepi_s6_2mm_66sl_PA_TR1.0_SBRef 29+cmrr_mbepi_s6_2mm_66sl_PA_TR1.0
15+cmrr_mbepi_s6_2mm_66sl_PA_TR1.0 99+PhoenixDocument
# Actually run the script...
> python ../Code/prisma_preproc.py -subject wl_subj046 \
-datadir "$PWD" \
-outdir ../Preproc \
-epis 15 17 19 21 23 25 27 29 \
-sbref 14 \
-distortPE 12 \
-distortrevPE 11 \
&> ../Preproc/prisma_preproc.log
Estimated Runtime: 1-8 hours; this step will require several hours to run, depending on the number of EPIs, their length, their resolution, as well as your computing power and memory limits.
This last command will produce a significant amount of output, all redirected to the
prisma_preproc.log file in the session’s Preproc directory. To understand the choice of
paramters, see the wiki page
documenting the script or the script’s help-text (python prisma_preproc.ph -h).
When this script has finished running, we can look at the results in the Preproc directory. Errors
that arise during the preprocessing script are documented on the wiki
page; to check for errors, use tail
prisma_preproc.log or less prisma_preproc.log to review the output from the script.
> cd ../Preproc
> ls
distort2anat_tkreg.dat timeseries_corrected_run03.nii.gz
distortion_merged_corrected.nii.gz timeseries_corrected_run04.nii.gz
distortion_merged_corrected_mean.nii.gz timeseries_corrected_run05.nii.gz
prisma_preproc.log timeseries_corrected_run06.nii.gz
sbref_reg_corrected.nii.gz timeseries_corrected_run07.nii.gz
session.json timeseries_corrected_run08.nii.gz
timeseries_corrected_run01.nii.gz workflow
timeseries_corrected_run02.nii.gz
> tail prisma_preproc.log
sub: /Volumes/server/Projects/Retinotopy/wl_subj046/20180205_ColorRetinotopy/Preproc/_merge_epis4/timeseries_corrected.nii.gz -> /Volumes/server/Projects/Retinotopy/wl_subj046/20180205_ColorRetinotopy/Preproc/timeseries_corrected_run05.nii.gz
180207-10:52:35,118 interface INFO:
sub: /Volumes/server/Projects/Retinotopy/wl_subj046/20180205_ColorRetinotopy/Preproc/_merge_epis5/timeseries_corrected.nii.gz -> /Volumes/server/Projects/Retinotopy/wl_subj046/20180205_ColorRetinotopy/Preproc/timeseries_corrected_run06.nii.gz
180207-10:52:42,585 interface INFO:
sub: /Volumes/server/Projects/Retinotopy/wl_subj046/20180205_ColorRetinotopy/Preproc/_merge_epis6/timeseries_corrected.nii.gz -> /Volumes/server/Projects/Retinotopy/wl_subj046/20180205_ColorRetinotopy/Preproc/timeseries_corrected_run07.nii.gz
180207-10:52:49,510 interface INFO:
sub: /Volumes/server/Projects/Retinotopy/wl_subj046/20180205_ColorRetinotopy/Preproc/_merge_epis7/timeseries_corrected.nii.gz -> /Volumes/server/Projects/Retinotopy/wl_subj046/20180205_ColorRetinotopy/Preproc/timeseries_corrected_run08.nii.gz
180207-10:52:58,230 workflow INFO:
[Job finished] jobname: outfiles jobid: 11
180207-10:52:58,237 workflow INFO:
Currently running 0 tasks, and 0 jobs ready. Free memory (GB): 43.20/43.20, Free processors: 8/8
As you can see, the end of the prisma_preproc.log does not contain any error messages; this is a
good sign and indicates that the script probably ran fine. In addition to the log file, we can see
that there are several timeseries_corrected_run*.nii.gz files containing the unwarped
motion-corrected time-series data for each EPI. Most of the other files can be ignored with the
exception of the distort2anat_tkreg.dat file, which contains the affine transform that
FreeSurfer can use to align the corrected timeseries data to the subject’s FreeSurfer anatomical
data. Warning: this file is not what it seems–specifically, it is not a simple
transformation from the affine-matrix stored in the time-series files to the subject’s FreeSurfer
anatomy (see next section). This alignment can be performed by the to_freesurfer.py script.
to_freesurfer.py
The to_freesurfer.py can be found in the same directory and location as the prisma_preproc.py
script. The purpose of the script is to apply the distort2anat_tkreg.dat alignment file to the
timeseries files in the Preproc directory after running the prisma_preproc.py script and to
create resampled cortical-surface timeseries files from the timeseries volume files; these can be
used to analyze the time-sereis data on the cortical surface directly.
Note: If you are using VistaSoft to analyze your pRF data (as done below in this document), then you do not need to perform this step. The Matlab scripts used to process the timeseries data will perform the realignment automatically.
The first step in the to_freesurfer.py script is to apply the distort2anat_tkreg.dat
transformation. Note that this file contains a tkreg transform, which is strikingly unintuitive if
you are not used to FreeSurfer’s way of thinking about MRI volumes. Typically, we assume that an
alignment transformation that aligns image A to image B would store that alignment as a
transformation from image A’s orientation to image B’s orientation: i.e., from affine matrix
would tell us how to change A’s affine transorm to align it to B. In a tkreg alignment matrix,
however, the transformation that is stored aligns A’s “tkreg” matrix to B.
In FreeSurfer, every MR image file has, in addition to any affine transformation that it stores, a
vox2ras-tkr (or “tkreg”) matrix. This matrix can be deduced from other header information, and can
be printed using the mri_info command:
> mri_info --vox2ras-tkr ./timeseries_corrected_run01.nii.gz
-2.00000 0.00000 0.00000 104.00000
0.00000 0.00000 2.00000 -66.00005
0.00000 -2.00000 0.00000 104.00000
0.00000 0.00000 0.00000 1.00000
This matrix is not the same as the image’s vox2ras or affine matrices; rather it is deduced
from the voxel size, image dimensions, and slice ordering. Notably, it does not change when the
affine transformation encoded in the file changes. Because this matrix is somewhat unintuitive, I
recommend applying the distort2anat_tkreg.dat transformation using the to_freesurfer.py script.
# to_freesurfer.py includes various options:
> python ../Code/to_freesurfer.py --help
usage: to_freesurfer.py [-h] [-t TAG] [-s] [-o OUTDIR] [-m METHOD] [-l LAYER]
[-d SDIR] [-v]
registration_file EPI [EPI ...]
positional arguments:
registration_file The distort2anat_tkreg.dat or similar file: the
registration file, in FreeSurfer\'s tkreg format, to
apply to the EPIs.
EPI The EPI files to be converted to anatomical
orientation
optional arguments:
-h, --help show this help message and exit
-t TAG, --tag TAG A tag to append to the output filenames; if given as -
then overwrites original files. By default, this is
"_anat".
-s, --surf If provided, instructs the script to also produce
files of the time-series resampled on the cortical
surface.
-o OUTDIR, --out OUTDIR
The output directory to which the files should be
written; by default this is the current directory (.);
note that if this directory also contains the EPI
files and there is no tag given, then the EPIs will be
overwritten.
-m METHOD, --method METHOD
The method to use for volume-to-surface interpolation;
this may be nearest or linear; the default is linear.
-l LAYER, --layer LAYER
Specifies the cortical layer to user in interpolation
from volume to surface. By default, uses midgray. May
be set to a value between 0 (white) and 1 (pial) to
specify an intermediate surface or may be simply
white, pial, or midgray.
-d SDIR, --subjects-dir SDIR
Specifies the subjects directory to use; by default
uses the environment variable SUBJECTS_DIR.
-v, --verbose Print verbose output
# (Optional) To preserve the original transformations, create a new directory
> mkdir ../Preproc_FreeSurfer
> cp ./distort2anat_tkreg.dat ./timeseries*.nii.gz ../Preproc_FreeSurfer
> cd ../Preproc_FreeSurfer
# It is typically run like this; this will only correct the timeseries headers
> python ../Code/to_freesurfer.py -v ./distort2anat_tkreg.dat timeseries*.nii.gz
Processing EPI ./timeseries_corrected_run01.nii.gz...
- Correcting volume orientation...
Processing EPI ./timeseries_corrected_run02.nii.gz...
- Correcting volume orientation...
Processing EPI ./timeseries_corrected_run03.nii.gz...
- Correcting volume orientation...
Processing EPI ./timeseries_corrected_run04.nii.gz...
- Correcting volume orientation...
Processing EPI ./timeseries_corrected_run05.nii.gz...
- Correcting volume orientation...
Processing EPI ./timeseries_corrected_run06.nii.gz...
- Correcting volume orientation...
Processing EPI ./timeseries_corrected_run07.nii.gz...
- Correcting volume orientation...
Processing EPI ./timeseries_corrected_run08.nii.gz...
- Correcting volume orientation...
# To also produce surface data-files
> python ../Code/to_freesurfer.py -v -s ./distort2anat_tkreg.dat timeseries*.nii.gz
Processing EPI ./timeseries_corrected_run01.nii.gz...
- Correcting volume orientation...
- Projecting to surface...
Processing EPI ./timeseries_corrected_run02.nii.gz...
- Correcting volume orientation...
- Projecting to surface...
Processing EPI ./timeseries_corrected_run03.nii.gz...
- Correcting volume orientation...
- Projecting to surface...
Processing EPI ./timeseries_corrected_run04.nii.gz...
- Correcting volume orientation...
- Projecting to surface...
Processing EPI ./timeseries_corrected_run05.nii.gz...
- Correcting volume orientation...
- Projecting to surface...
Processing EPI ./timeseries_corrected_run06.nii.gz...
- Correcting volume orientation...
- Projecting to surface...
Processing EPI ./timeseries_corrected_run07.nii.gz...
- Correcting volume orientation...
- Projecting to surface...
Processing EPI ./timeseries_corrected_run08.nii.gz...
- Correcting volume orientation...
- Projecting to surface...
> ls
distort2anat_tkreg.dat rh.timeseries_corrected_run06.mgz
lh.timeseries_corrected_run01.mgz rh.timeseries_corrected_run07.mgz
lh.timeseries_corrected_run02.mgz rh.timeseries_corrected_run08.mgz
lh.timeseries_corrected_run03.mgz timeseries_corrected_run01.nii.gz
lh.timeseries_corrected_run04.mgz timeseries_corrected_run02.nii.gz
lh.timeseries_corrected_run05.mgz timeseries_corrected_run03.nii.gz
lh.timeseries_corrected_run06.mgz timeseries_corrected_run04.nii.gz
lh.timeseries_corrected_run07.mgz timeseries_corrected_run05.nii.gz
lh.timeseries_corrected_run08.mgz timeseries_corrected_run06.nii.gz
rh.timeseries_corrected_run01.mgz timeseries_corrected_run07.nii.gz
rh.timeseries_corrected_run02.mgz timeseries_corrected_run08.nii.gz
rh.timeseries_corrected_run03.mgz
rh.timeseries_corrected_run04.mgz
rh.timeseries_corrected_run05.mgz
Estimated Runtime: a couple minutes per EPI.
pRF Models
Solving the pRF models based on the timeseries data is performed using VistaSoft in Matlab. The three scripts documented below are designed to be run on the Winawer-lab server using the directory structures shown in this document. For deviations from this narrow use-case, you will need to edit the scripts themselves. Mostly, these edits can probably be small (e.g., if you are using a different set of directory names), but for different pipelines, these scripts should be seen as a rough guide and not a solution. In these cases, I would recomment the VistaSoft Ernie tutorials and related VistaSoft tutorials on MRI data analysis.
All of these scripts can be found in the MRI_tools
repository in the retinotopy directory. They require that the
ToolboxToolbox for Matlab be installed; this will
manage other dependencies, including VistaSoft. Recall that we already copied all of these scripts
into our session’s Code directory when we setup the session directory. All three of these scripts
will deduce the subject, session, and directory structure when run as long as you are using a
similar structure as this set of demos. You can also edit the top parts of the scripts
init_vista.m
The init_vista script initializes the subject’s VistaSoft session; this primarily involves
initializing certain data structures, finding the preprocessed timeseries files, and applying the
appropriate alignments. If the subject does not yet have a VistaSoft anatomy directory in the
/Volumes/server/Projects/Anatomy directory (where we store VistaSoft anatomical sessions in the
Winawer lab), it will create one of these and initialize it from the subject’s FreeSurfer
directory.
To run init_vista, simply invoke it from inside your session’s Code directory:
>> cd('/Volumes/server/Projects/Retinotopy/wl_subj046/20180205_ColorRetinotopy/Code');
>> init_vista
%% (This command generates significant output that is not reproduced here.)
Estimated Runtime: fast, usually less than a minute; could stretch up to 10 minutes if it has to initialize the Anatomy session and your computer is fairly slow.
solve_pRFs.m
The solve_pRFs script calculates the actual solutions to pRF models for all of the subject’s
gray-matter voxels using the timeseries data. If you don’t edit the script, this creates a single
dataset named ‘Full’, which contains the average of all the timeseries data, and solves only this
dataset. If you wish to analyze separate subsets of the data, you must edit this script. That said,
adding new datasets is trivial: simply search your local copy of the script for #scan_plan and
read the comment containing it.
To run solve_pRFs, simply invoke it from inside your session’s Code directory:
>> cd('/Volumes/server/Projects/Retinotopy/wl_subj046/20180205_ColorRetinotopy/Code');
>> solve_pRFs
%% (This command generates significant output that is not reproduced here.)
export_niftis.m
The export_niftis script writes the pRF model solutions out to a set of nifti files in your
session’s Outputs directory. As with solve_pRFs, the default behavior is to export a single
‘Full’ dataset; to change this, you will need to edit the script. As with solve_pRFs, this is made
easy by instructions in a comment tagged with #output_plan.
To run export_niftis, simply invoke it from inside your session’s Code directory:
>> cd('/Volumes/server/Projects/Retinotopy/wl_subj046/20180205_ColorRetinotopy/Code');
>> export_niftis
%% (This command generates significant output that is not reproduced here.)
The files output by export_niftis contain the following data (here, as if the output prefix were
‘full’, which is also the default behavior):
full-xcrds.nii.gzandfull-ycrds.nii.gzcontain the coordinates of the pRF centers in degrees of the visual field.full-vexpl.nii.gzcontains the proportion of variance explained by the model (a fraction between 0 and 1).full-sigma.nii.gzcontains the effective pRF radius in degrees of the visual field.
postproc_pRFs.py
The final script in the retinotopy directory of the MRI_tools repository is the
postproc_pRFs.py
script. This script reads in the files exported by the export_niftis.m script and creates volume
files for polar angle and eccentricity as well as surface files for all data types (polar angle,
eccentricity, x, y, variance explained, sigma). The script requires merely the FreeSurfer subject id
and the path to the Outputs directory. It automatically operates on all datasets found there.
> pwd
/Volumes/server/Projects/Retinotopy/wl_subj046/20180205_ColorRetinotopy
> ls Outputs
all-sigma.nii.gz all-vexpl.nii.gz all-xcrds.nii.gz all-ycrds.nii.gz
> python ~/Code/MRI_tools/retinotopy/postproc_pRFs.py wl_subj046 Outputs -v
Processing Dataset all...
- Importing parameters...
- Creating polar angle/eccentricity images...
- Projecting to surface...
> ls Outputs
all-angle.nii.gz all-xcrds.nii.gz lh.all-sigma.mgz rh.all-angle.mgz rh.all-xcrds.mgz
all-eccen.nii.gz all-ycrds.nii.gz lh.all-vexpl.mgz rh.all-eccen.mgz rh.all-ycrds.mgz
all-sigma.nii.gz lh.all-angle.mgz lh.all-xcrds.mgz rh.all-sigma.mgz
all-vexpl.nii.gz lh.all-eccen.mgz lh.all-ycrds.mgz rh.all-vexpl.mgz
Surfaces and Parameter Visualization
A simple way to check that the above steps ran correctly is to visualize the solved pRF parameters projected to the cortical surface; this essentially represents the culmination of all of the steps documented so far in these tutorials. FreeView is capable of performing this kind of visualization, as well as various other tools and libraries. For simple checks, FreeView is sufficient; for more complex visualization, I would suggest a combination of neuropythy and pycortex, or Neurotica if you use Mathematica.
To start FreeView, we want to load the subject’s inflated hemisphere, which makes visualization easiest; alternately, loading the pial or white hemispheres can be useful for orienting oneself relative to anatomical landmarks. Here we will look at only one hemisphere, but examining the other hemisphere should be straightforward given the commands for one.
# Open FreeView with the subject's LH inflated surface
> freeview -f "$SUBJECTS_DIR"/wl_subj046/surf/lh.inflated
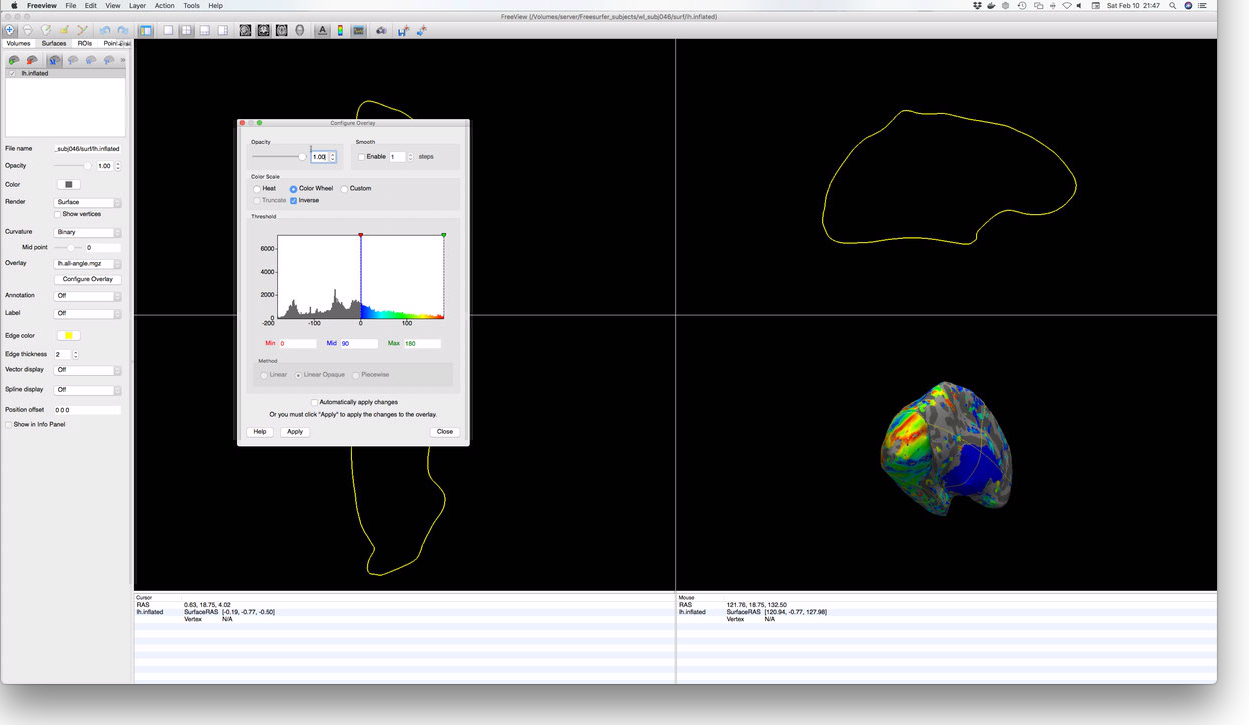
This should open FreeView with the inflated left hemisphere shown in the lower right display
window. To add an overlay, navigate to the Surfaces tab in the upper left of the FreeView
window. When you click on Surfaces, the left menu will change; among the surface controls revealed
is a dropdown menu labeled ‘Overlay’. One of the options in this list is ‘Load generic overlay…’.
Selecting this item will allow you to navigate to your subject’s Outputs directory to load the
lh.all-angle.mgz file, containing the polar-angle measurements for the subject’s left
hemisphere. Once the overlay is loaded, it will likely need to be configured via the Configure
Overlay button. These controls are not perfectly intuitive, but they are not difficult to figure
out by trial and error. When properly configured, you should see something like this:
Atlas Generation
Anatomical atlases are a common and useful way to examine how your subject compares with average
retinotopic mapping data. The atlases described here use cortical surface normalization (i.e., via
FreeSurfer’s fsaverage or fsaverage_sym subjects) in order to describe the average expected
location of various ROIs and/or their average expected organizational properties. Here we will look
at two atlases:
- The “Benson-2014” atlas: Benson NC, Butt OH, Brainard DH, Aguirre GK (2014) Correction of Distortion in Flattened Representations of the Cortical Surface Allows Prediction of V1-V3 Functional Organization from Anatomy. PLOS Comput Biol 10(3):e1003538. This atlas describes the location and retinotopic organization of V1, V2, and V3, as well as several higher areas (though these areas are not considered authoritative by the authors as they have not been tested). Note that the Benson-2014 atlas predicts retinotopic maps from 0-90 degrees of eccentricity, though only the inner 20 degrees of eccentricity in V1, V2, and V3 has been validated. This atlas provides visual area label, polar angle, eccentricity, and pRF radius (sigma) predictions.
- The “Wang-2015” atlas: Wang L, Mruczek RE, Arcaro MJ, Kastner S (2015) Probabilistic Maps of Visual Topography in Human Cortex. Cereb Cortex 25(10):3911-31. This atlas describes the locations/boundaries of 25 visual areas with emphasis on dorsal visual areas. This atlas does not describe retinotopy but only average expected ROI labels.
Wang2015 Atlas
The easiest way to apply the Wang-2015 atlas is to use the occipital_atlas docker, information
about which can be found here. See this link for
documentation on using the Docker.
Benson2014 Atlas
The easiest way to apply the Benson-2014 atlas is to, again, use
neuropythy. The library includes a builtin command,
benson14_retinotopy that places the atlas files in your subject’s surf/ and mri/
directories–surface files in the surf/ directory and volumetric images in the mri/
directory. It can be executed like so:
> python -m neuropythy benson14_retinotopy wl_subj046 -v
Processing subject wl_subj046:
- Interpolating template...
- Exporting surfaces:
- Exporting LH prediction file: /Users/nben/Documents/WinawerLab/Freesurfer_subjects/wl_subj046/surf/lh.benson14_varea
- Exporting LH prediction file: /Users/nben/Documents/WinawerLab/Freesurfer_subjects/wl_subj046/surf/lh.benson14_eccen
- Exporting LH prediction file: /Users/nben/Documents/WinawerLab/Freesurfer_subjects/wl_subj046/surf/lh.benson14_angle
- Exporting LH prediction file: /Users/nben/Documents/WinawerLab/Freesurfer_subjects/wl_subj046/surf/lh.benson14_sigma
- Exporting RH prediction file: /Users/nben/Documents/WinawerLab/Freesurfer_subjects/wl_subj046/surf/rh.benson14_varea
- Exporting RH prediction file: /Users/nben/Documents/WinawerLab/Freesurfer_subjects/wl_subj046/surf/rh.benson14_eccen
- Exporting RH prediction file: /Users/nben/Documents/WinawerLab/Freesurfer_subjects/wl_subj046/surf/rh.benson14_angle
- Exporting RH prediction file: /Users/nben/Documents/WinawerLab/Freesurfer_subjects/wl_subj046/surf/rh.benson14_sigma
- Exporting Volumes:
- Preparing volume file: /Users/nben/Documents/WinawerLab/Freesurfer_subjects/wl_subj046/mri/benson14_varea.mgz
- Exporting volume file: /Users/nben/Documents/WinawerLab/Freesurfer_subjects/wl_subj046/mri/benson14_varea.mgz
- Preparing volume file: /Users/nben/Documents/WinawerLab/Freesurfer_subjects/wl_subj046/mri/benson14_eccen.mgz
- Exporting volume file: /Users/nben/Documents/WinawerLab/Freesurfer_subjects/wl_subj046/mri/benson14_eccen.mgz
- Preparing volume file: /Users/nben/Documents/WinawerLab/Freesurfer_subjects/wl_subj046/mri/benson14_angle.mgz
- Exporting volume file: /Users/nben/Documents/WinawerLab/Freesurfer_subjects/wl_subj046/mri/benson14_angle.mgz
- Preparing volume file: /Users/nben/Documents/WinawerLab/Freesurfer_subjects/wl_subj046/mri/benson14_sigma.mgz
- Exporting volume file: /Users/nben/Documents/WinawerLab/Freesurfer_subjects/wl_subj046/mri/benson14_sigma.mgz
Subject wl_subj046 finished!
> ls "$SUBJECTS_DIR"/wl_subj046/*/*benson14*
/Volumes/server/Freesurfer_subjects/wl_subj046/mri/benson14_angle.mgz
/Volumes/server/Freesurfer_subjects/wl_subj046/mri/benson14_eccen.mgz
/Volumes/server/Freesurfer_subjects/wl_subj046/mri/benson14_sigma.mgz
/Volumes/server/Freesurfer_subjects/wl_subj046/mri/benson14_varea.mgz
/Volumes/server/Freesurfer_subjects/wl_subj046/surf/lh.benson14_angle
/Volumes/server/Freesurfer_subjects/wl_subj046/surf/lh.benson14_eccen
/Volumes/server/Freesurfer_subjects/wl_subj046/surf/lh.benson14_sigma
/Volumes/server/Freesurfer_subjects/wl_subj046/surf/lh.benson14_varea
/Volumes/server/Freesurfer_subjects/wl_subj046/surf/rh.benson14_angle
/Volumes/server/Freesurfer_subjects/wl_subj046/surf/rh.benson14_eccen
/Volumes/server/Freesurfer_subjects/wl_subj046/surf/rh.benson14_sigma
/Volumes/server/Freesurfer_subjects/wl_subj046/surf/rh.benson14_varea
Estimated Runtime: 20 minutes or less; this step is not terribly intensive, but template interpolation can take awhile on a slower computer.
Note that the surface-file outputs of this command are in FreeSurfer’s ‘curv’ format (morph_data
if you are using nibabel). If you prefer mgz format, you can use the --surf-format=mgz option;
see python -m neuropythy benson14_retinotopy --help for more information. Similarly, the volume
files may be written as .nii.gz nifti files instead of .mgz files via the argument
--vol-format=nifti.
Bayesian Inference of Retinotopic Maps
Performing Bayesian inference on the retinotopic mapping data in order to generate an inferred map
prediction is as simple as a single command (albeit one with several arguments) executed by the
neuropythy library. The following code-block
demonstrates this. After running the command, files named similar to lh.inferred_angle.mgz
describe the predicted polar angle, eccentricity, visual area (varea) and pRF radius (sigma).
> pwd
/Volumes/server/Projects/Retinotopy/wl_subj046/20180205_ColorRetinotopy/Outputs
> ls
all-angle.nii.gz all-xcrds.nii.gz lh.all-sigma.mgz rh.all-angle.mgz rh.all-xcrds.mgz
all-eccen.nii.gz all-ycrds.nii.gz lh.all-vexpl.mgz rh.all-eccen.mgz rh.all-ycrds.mgz
all-sigma.nii.gz lh.all-angle.mgz lh.all-xcrds.mgz rh.all-sigma.mgz
all-vexpl.nii.gz lh.all-eccen.mgz lh.all-ycrds.mgz rh.all-vexpl.mgz
> python -m neuropythy register_retinotopy wl_subj046 \
--verbose \
--surf-outdir=. --surf-format="mgz" \
--no-volume-export \
--lh-angle=lh.all-angle.mgz \
--lh-eccen=lh.all-eccen.mgz \
--lh-weight=lh.all-vexpl.mgz \
--lh-radius=lh.all-sigma.mgz \
--rh-angle=rh.all-angle.mgz \
--rh-eccen=rh.all-eccen.mgz \
--rh-weight=rh.all-vexpl.mgz \
--rh-radius=rh.all-sigma.mgz
Processing subject: wl_subj046
Preparing RH Registration...
Preparing LH Registration...
Exporting files...
Extracting RH predicted mesh...
Extracting LH predicted mesh...
# This command produces some new files:
> ls
all-angle.nii.gz lh.all-xcrds.mgz rh.all-vexpl.mgz
all-eccen.nii.gz lh.all-ycrds.mgz rh.all-xcrds.mgz
all-sigma.nii.gz lh.inferred_angle.mgz rh.all-ycrds.mgz
all-vexpl.nii.gz lh.inferred_eccen.mgz rh.inferred_angle.mgz
all-xcrds.nii.gz lh.inferred_sigma.mgz rh.inferred_eccen.mgz
all-ycrds.nii.gz lh.inferred_varea.mgz rh.inferred_sigma.mgz
lh.all-angle.mgz lh.retinotopy.sphere.reg rh.inferred_varea.mgz
lh.all-eccen.mgz rh.all-angle.mgz rh.retinotopy.sphere.reg
lh.all-sigma.mgz rh.all-eccen.mgz
lh.all-vexpl.mgz rh.all-sigma.mgz
Estimated Runtime: Variable, but if you stick to the default number of steps and have a reasonably powerful computer, this should take less than an hour per hemisphere. If you have only one core, for example, this may take a few hours. With a powerful machine, this command can take as little as 15 minutes per hemisphere.